Cytoplasm Subsystem: Difference between revisions
| Line 28: | Line 28: | ||
The simplest implementation of transcription and translation makes use of the pre-defined `TxTlExtract` mixt | The simplest implementation of transcription and translation makes use of the pre-defined `TxTlExtract` mixt | ||
<pre> | |||
import numpy as np | |||
import matplotlib.pyplot as plt | |||
import biocrnpyler as bcp | |||
# Create a simple GFP DNA construct | |||
gfp_DNA = bcp.DNAassembly('GFP', promoter='pconst', rbs='rbs') | |||
# Create a TXTL mixture containing the GFP | |||
mixture = bcp.TxTlExtract( | |||
"TX-TL of GFP", components=[gfp_DNA], | |||
parameters={'ktx': 0.5, 'ktl': 2, 'kb': 100, 'ku':10, 'kdeg': 100}) | |||
# Compile the CRN and generate a simulation | |||
gfp_CRN = mixture.compile_crn() | |||
timepts = np.linspace(0, 100) | |||
gfp_sim = gfp_CRN.simulate_with_bioscrape_via_sbml( | |||
timepts, initial_condition_dict={ | |||
'dna_GFP': 1, 'protein_RNAP': 10, 'protein_Ribo': 50}) | |||
plt.plot(timepts, gfp_sim['protein_GFP']) | |||
plt.xlabel("Time, min") # default units = minutes | |||
plt.ylabel("[GFP], nM") # default units = nM | |||
</pre> | |||
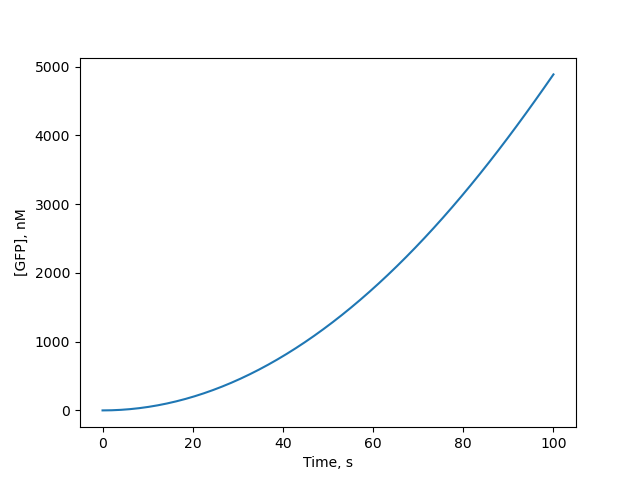
yielding the plot | yielding the plot | ||
[[Image:gfp_sim-plot.png]] | [[Image:gfp_sim-plot.png]] | ||
Revision as of 07:31, 7 July 2025
The "cytoplasm" subsystem is responsible for implementing and maintaining the internal environment of the synthetic cell, including key mechanisms such as transcription, translation, and degradation. There are many different approaches to implementing the cytoplasm subsystem, depending on the desired functionality of the synthetic cell. On this page we describe some of the key functions and approaches to implementing those functions.
Transcription and Translation
The functions of transcription and translation enable to expression of RNA and proteins from DNA. They are often key elements in synthetic cells since they allow genetic programs that can conditionally express molecules with prescribed functionality.
Transcription only systems
Systems that implement just transcription, such as genelets.
Purified transcription and translation systems (PURE)
PURExpress, PUREFrex, OnePot pure
Extract-based transcription and translation systems (TX-TL)
Degradation and Waste Management
Another function of the cytoplasm subsystem is to help degrade products of transcription and translation when they are no longer needed and to manage any waste products that are produced by the cytoplasm. These functions often make use of other subsystems, such as the Transport Subsystem.
Degradation tags
External buffers
Non-Transcription/Translation-Based Systems
In some cases, a synthetic cell may not make use of transcription nor translation as its mechanism for implementing the functionality of synthetic cells.
Genelets
Implementation in BioCRNpyler
BioCRNpyler contains a number of modules and mechanisms that can be used to model the cytoplasm subsystem.
The simplest implementation of transcription and translation makes use of the pre-defined `TxTlExtract` mixt
import numpy as np
import matplotlib.pyplot as plt
import biocrnpyler as bcp
# Create a simple GFP DNA construct
gfp_DNA = bcp.DNAassembly('GFP', promoter='pconst', rbs='rbs')
# Create a TXTL mixture containing the GFP
mixture = bcp.TxTlExtract(
"TX-TL of GFP", components=[gfp_DNA],
parameters={'ktx': 0.5, 'ktl': 2, 'kb': 100, 'ku':10, 'kdeg': 100})
# Compile the CRN and generate a simulation
gfp_CRN = mixture.compile_crn()
timepts = np.linspace(0, 100)
gfp_sim = gfp_CRN.simulate_with_bioscrape_via_sbml(
timepts, initial_condition_dict={
'dna_GFP': 1, 'protein_RNAP': 10, 'protein_Ribo': 50})
plt.plot(timepts, gfp_sim['protein_GFP'])
plt.xlabel("Time, min") # default units = minutes
plt.ylabel("[GFP], nM") # default units = nM
yielding the plot